tRForest
Explore machine-learning predicted targets
for Transfer RNA-related fragments (tRFs)


Explore machine-learning predicted targets
for Transfer RNA-related fragments (tRFs)



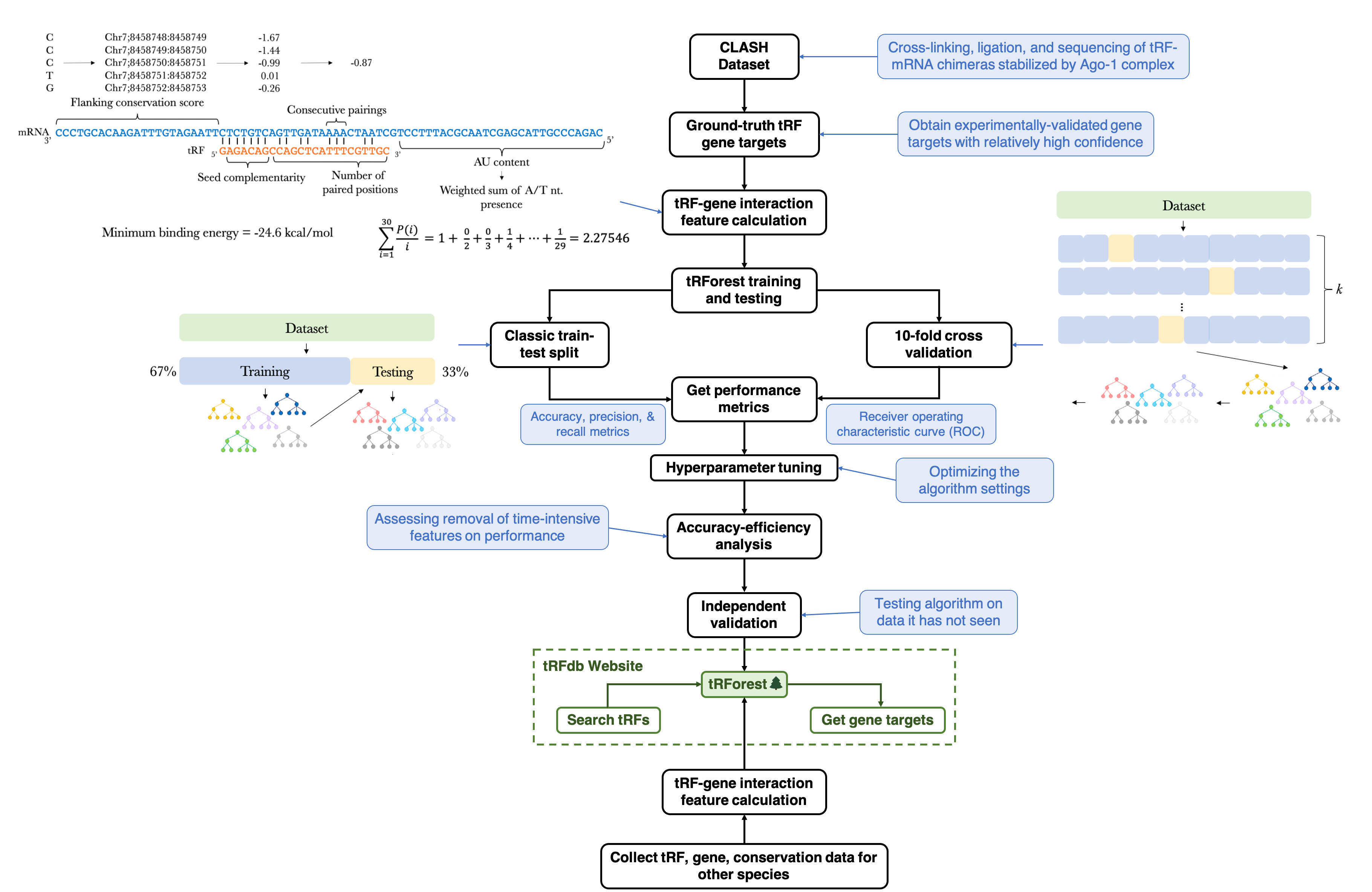
tRF IDs and tRF sequences for all species were obtained from tRFdb. Gene names, transcript IDs, 3' UTR sequences, and chromosomal coordinates for all species were obtained from Ensembl Biomart. The CLASH dataset used to obtain ground-truth tRF targets can be downloaded here. Base-wise phyloP conservation score mapping files were obtained from the UCSC Genome Browser.
RNAduplex and RNAfold from the ViennaRNA package were used to determine binding location/energy and energy cutoffs for tRF-target duplexes. Bedops was used to calculate two features, stem and flanking conservation scores, for tRF-target duplexes. RNAhybrid was used to create the interaction illustrations. The sci-kit learn package was used to create the tRForest machine-learning algorithm.